
Designing Drug Candidates with Generative Adversarial Networks
This project tackles the challenges of traditional drug discovery, which is often a slow, expensive, and inefficient process. Faced with a vast chemical space of up to (10^{60}) potential molecules, this research proposes a computational approach using deep learning to accelerate the design of new drug candidates. The core of this project is a generative model designed to produce novel, valid, and optimized molecules represented in SMILES (Simplified Molecular Input Line Entry Strings) format.
Model Architecture
Model Diagram:
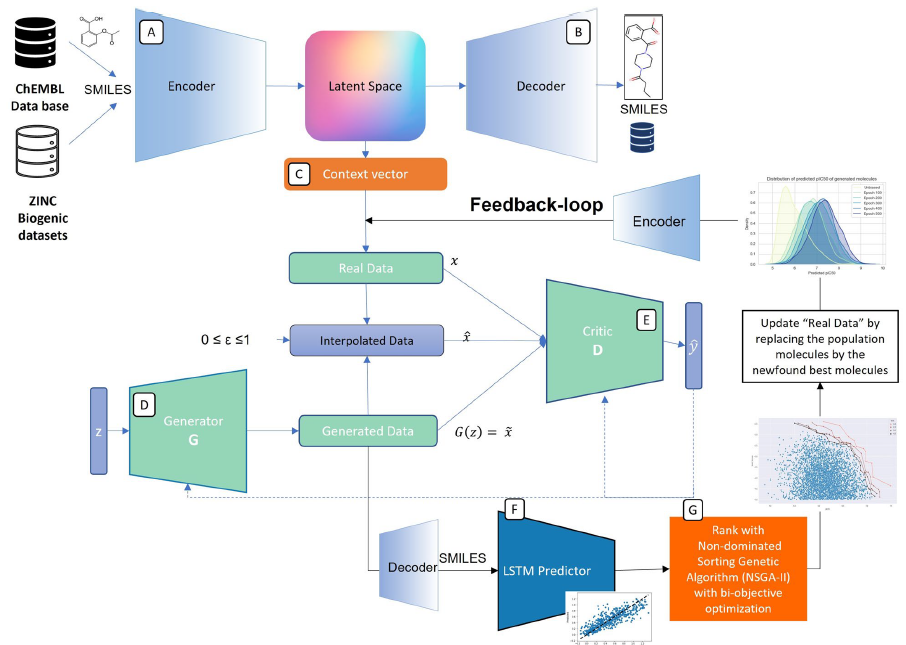
VAE-WGAN-GP
The model combines a Variational Autoencoder (VAE) with a Wasserstein Generative Adversarial Network with Gradient Penalty (WGAN-GP). This hybrid architecture leverages the VAE's ability to represent discrete molecular structures in a continuous space, which is ideal for training the GAN.
-
Input:
- The model is trained on a dataset of 100,000 SMILES strings extracted from the ChEMBL and Zinc Biogenic public databases.
- The generator component takes a random noise vector of size 256 as its initial input.
-
Preprocessing and VAE Encoder:
- SMILES strings are first preprocessed through standardization, tokenization, and padding.
- The VAE's Encoder then transforms these strings into 256-dimensional continuous latent vectors, which serve as the "real" data for the GAN training.
-
WGAN-GP Core:
- The Generator network attempts to produce realistic 256-dimensional latent vectors from the initial random noise.
- The Critic (Discriminator) is trained to distinguish between the real latent vectors (from the Encoder) and the fake ones (from the Generator). It uses a Wasserstein loss function with a gradient penalty to ensure training stability and avoid issues like vanishing gradients.
-
Decoder and Output:
- Once the GAN is trained, the Generator can produce new, optimized latent vectors.
- These generated vectors are passed to the VAE's Decoder, which converts them back into novel SMILES strings, representing new potential drug candidates.
Results
The model was evaluated by generating 100 new molecules and assessing them on several key criteria. The primary evaluation metrics were Validity, logP (partition coefficient), and Molecular Weight. Quantitative analysis showed that the model's ability to generate valid molecules progressively increased with more training epochs. The generated molecules were further analyzed for their drug-like properties (logP and molecular weight) to determine their potential as effective drug candidates.
Explanation of Molecular Validity:
A Valid Molecule refers to a generated SMILES string that represents a chemically correct and synthesizable structure. In this project, validity was confirmed using the RDKit software library. This is the most critical initial metric because it ensures that the model is not producing nonsensical or physically impossible chemical structures, making the generated candidates viable for further testing and consideration.